Chemistry Department
Hamilton College
198 College Hill Road
Clinton, NY 13323
Adam Van Wynsberghe, Physical Chemistry

Associate Professor
Office: Taylor Science Center 1063
Phone: 315-859-4309
Email: avanwyns@hamilton.edu
At Hamilton since 2009
Van Wynsberghe Lab Web Page >>
EDUCATION
B.A. Ohio Wesleyan University, Biochemistry; Minor: Mathematics, 2001.
Ph.D. University of Wisconsin-Madison with Qiang Cui, Biophysics, 2007.
NIH NRSA Post-Doctoral Fellow at UC-San Diego with J. Andrew McCammon, 2007-2009.
RESEARCH
Our lab uses theoretical and computational techniques to investigate the interactions and dynamics of biological macromolecules. We hope to help elucidate the mechanisms by which these marcromolecules utilize intramolecular motion and intermolecular forces to achieve their biological functions. Currently, our main focus has been on exploring the pathways and kinetics of ligands binding to the influenza protein neuraminidase (Figure 1).
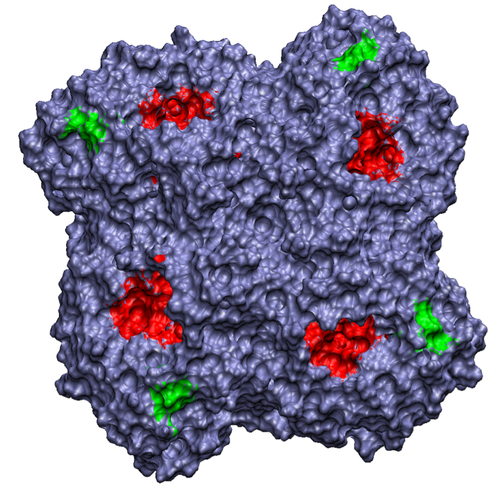
Figure 1. Influenza neuraminidase tetramer; active sites
are shown in red, secondary sites are shown in green.
We are currently developing a computational methodology to accurately measure the association rate constants of small ligands to both the active (red in Figure 1) and secondary (green in Figure 1) binding sites. This methodology models the diffusional character of the association pathway using Brownian dynamics (BD) and the local, van der Waal contact search with classical molecular dynamics (MD) simulations. As shown in Figure 2, we are currently attempting to implement this multi-scale method by switching from the approximate BD to the more detailed MD at a proximal elliptical surface. The qualitative application of this methodology has allowed us to computationally predict the pathways of various ligands binding to neuraminidases from a variety of sources.
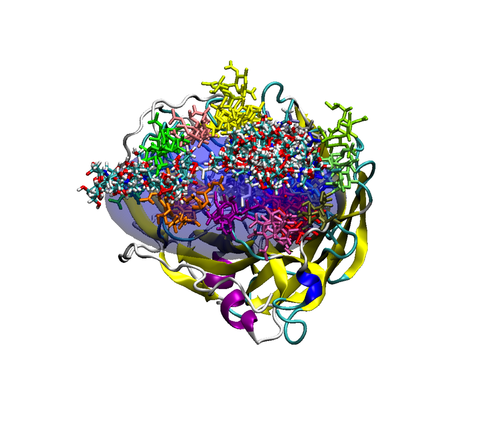
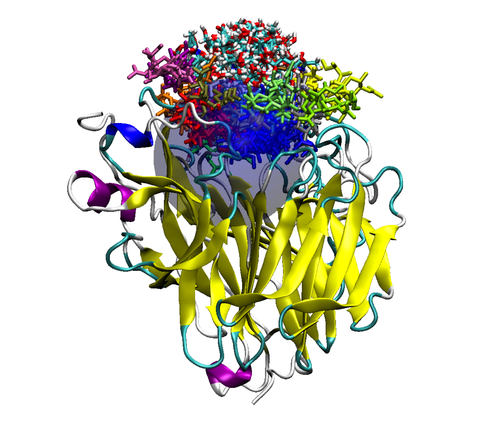
Figure 2. Top (left panel) and side (right panel) view of a neuraminidase monomer. The elliptical surface to switch between BD and MD is shown as partially
transparent. Ligands in atom-type coloring are BD arrivals to the transition surface; ligands in single colors are MD bound to the surface of the protein.
COURSES TAUGHT
Chemistry 321: Physical Chemistry I
Chemistry 321L: Physical Chemistry I Laboratory
Chemistry 322: Physical Chemistry II
Chemistry 322L: Physical Chemistry II Laboratory
Chemistry 120: Principles of Chemistry
Chemistry 120L: Principles of Chemistry Laboratory
Chemistry 125: Principles to Applications
Chemistry 125L: Principles to Applications Laboratory
Chemistry 270L Biological Chemistry Laboratory
GRANTS
National Science Foundation: Major Research Instrumentation, $225,000, 2016-2019, Contributor, MRI: Addition of High Performance Computers for the Molecular Education and Research Consortium in Undergraduate computational chemistRY (MERCURY).
Hamilton College: Class of 1963 Faculty Fellowship, $3,500, 2016, PI, Development of a water quality theme for Chemistry 125.
Hamilton College: Class of 1966 Career Development Award, $4,800, 2015, PI, Development of BPS and PFOA Detection Methodology.
XSEDE Allocation: TG-MCB130192, 100,000 Service Units, 2013-2014, PI, A Study of Sialic Acid-Neuraminidase Binding Events through MM/PBSA Free-Energy Calculations. Co-Pi was Hamilton undergraduate Leah Krause '14
National Science Foundation: Major Research Instrumentation, $200,000, 2012-2015, Cobntirbutor, MRI: Acquisition of a High Performance Computer for the Molecular Education and Research Consortium in Undergraduate computational chemistRY (MERCURY)
Research Corporation: Cottrell College Science Award, $35,000 plus $30,000 Hamilton College match, 2010-2012, PI, Accurate Evaluation of Association Rate Constants of Influenza Neuraminidase Inhibitors.
National Science Foundation: Major Research Instrumentation, $177,950, 2010-2012, PI, MRI-R2: Acquisition of a High Performance Computing cluster with a fast interconnect to enable shared-use, college-wide computational investigations at Hamilton College.
Teragrid Allocation: TG-MCB090196, 200,000 Service Units, 2009-2010, PI, Investigations of Ribonuclease A Catalysis.
AWARDS
2016 Class of 1963 Faculty Fellowship
2015 Class of 1966 Career Development Award
2014 John R. Hatch, Class of 1925, Excellence in Teaching Award
2012 National Biomedical Computational Resource Summer Institute Scholarship
2008-2009 NIH (NIGMS) Ruth L. Kirschstein NRSA Post-Doctoral Fellowship
2003-2005 National Science Foundation Pre-Doctoral Fellowship
2001 Department of Defense Pre-Doctoral Fellowship (declined)
2001 Honorable Mention, Howard Hughes Medical Institute Pre-Doctoral Fellowship
2001-2002 University of Wisconsin-Madison WARF Prize Fellowship
2001 Slocum Prize in the Sciences (Highest Overall GPA), Ohio Wesleyan University
2001 Outstanding Senior Chemistry Major, Ohio Wesleyan University
2001 Phi Beta Kappa Honor Society, Ohio Wesleyan University
2000 Barry M. Goldwater Scholarship
2000 Ernest B. Yeager Award, Society for Applied Spectroscopy, Cleveland Section
2000 Outstanding Junior Chemistry Major, Ohio Wesleyan University
1999 Outstanding Poster, Central Ohio Undergraduate Research Symposium
1997 National Merit Finalist
PUBLICATIONS
(*denotes undergraduate co-author)
- O'Grady, C.E.*, P. Talpey*, T. E. Elgren, and A. W. Van Wynsberghe, 2014. The development and implementation of a bio-molecular docking exercise for the general chemistry laboratory. In Annual Reports in Computational Chemistry. Vol. 10. Ralph Wheeler, editor. Elsevier, United Kingdom, pp. 167-187.
- Kinnel, R. B., A. W. Van Wynsberghe, I. J. Rosenstein, K. S. Brewer, M. Cotten, G. C. Shields, C. J. Borton, S. Z. Senior, G. S. Rahn, and T. E. Elgren, 2013. A Departmental Focus on High Impact Undergraduate Research Experiences. In Developing and Maintaining a Successful Undergraduate Research Program. T. W. Chapp and M. A. Benvenuto, editors. ACS Symposium Series; American Chemical Society: Washington, D.C., vol. 1156:5-22.
- Tsutakawa, S. E., A. W. Van Wynsberghe, B. D. Freudenthal, C. P. Weinacht, L. Gakhar, M. T. Washington, Z. Zhuang, J. A. Tainer, and I. Ivanov, 2011. Solution X-ray scattering combined with computational modeling reveals multiple conformations of covalently-bound ubiquitin on PCNA. Proc. Natl. Acad. Sci. 108(43):17672-17677.
- Sinko, W., C. de Oliveira, S. Williams, A. Van Wynsberghe, J. Durrant, R. Cao, E. Oldfi eld, and J. A. McCammon, 2011. Applying Molecular Dynamics Simulations to Identify Rarely Sampled Ligand bound Conformational States of Undecaprenyl Pyrophosphate Synthase, an Antibacterial Target. Chem. Biol. Drug Des. 77(6):412-420.
- Van Wynsberghe, A. W. and Q. Cui, 2010. Conservation and Variation of Structural Flexibility in Protein Families. Structure. 18:281-283.
- Sung, J. C.*, A. W. Van Wynsberghe, R. E. Amaro, W. W. Li, and J. A. McCammon, 2010. The role of secondary sialic acid binding sites in influenza N1 neuraminidase. J. Amer. Chem. Soc. 132(9):2883-2885.
- Van Wynsberghe, A. W., L. Ma, X. Chen, and Q. Cui, 2008. Functional Motions in Biomolecules: Insights from Computational Studies at Multiple Scales. In Computational Structural Biology. T. Schwede and M. Peitsch, editors. World Scienti c Publishing.
- Kondrashov, D., A. Van Wynsberghe, R. M. Bannen, Q. Cui, and G. N. Phillips, Jr., 2007. Protein Structural Variation in Computational Models and Crystallographic Data. Structure. 15:169-177.
- Van Wynsberghe, A., and Q. Cui, 2006. Interpreting correlated motions using normal mode analysis. Structure. 14:1647-1653.
- Van Wynsberghe, A. W., and Q. Cui. 2005. Comparison of mode analyses at di fferent resolutions applied to nucleic acid systems. Biophys. J. 89:2939-2949.
- Li, G., A. Van Wynsberghe, O. N. A. Demerdash, and Q. Cui. 2005. Normal Mode Analysis of Macromolecules: From Enzyme Active Sites to Molecular Machines. In Normal Mode Analysis: Theory and Applications to Biological and Chemical Systems. Q. Cui and I. Bahar, editors. CRC Press, Boca Raton.
- Felitsky, D. J., J. G. Cannon, M. W. Capp, J. Hong, A. W. Van Wynsberghe, C. F. Anderson, and M. T. Record. 2004. The exclusion of glycine betaine from anionic biopolymer surface: Why glycine betaine is an e ective osmoprotectant but also a compatible solute. Biochem. 43:14732-14743.
- Van Wynsberghe, A., G. H. Li, and Q. Cui. 2004. Normal-mode analysis suggests protein flexibility modulation throughout RNA polymerase's functional cycle. Biochem. 43:13083-13096.
- Van Wynsberghe, A. W., S. A. Peebles, R. A. Peebles, and R. L. Kuczkowski. 2000. Rotational spectrum and structure of 1,2-dichloro-3,3,4,4-tetrafluorocyclobutene: Comparison of spectroscopy, di raction, and ab initio results. J. Phys. Chem. A 104:8702-8708.
RECENT STUDENT PRESENTATIONS
(*denotes undergraduate co-author)
- Kang, J* and A. W. Van Wynsberghe, 2017. Computational Investigation of the Binding Pathways of Zanamivir to H274Y Neuraminidase. Poster- 16th MERCURY Conference, Greenville, SC.
- Nash, K. L.*, R. W. Wenner*, and A. W. Van Wynsberghe, 2017. Exploration of the Binding Kinetics of Zanamivir to WT Neuraminidase via MM/GBSA Analysis of Molecular Dynamics Simulations. Poster- 16th MERCURY Conference, Greenville, SC.
- Park, A* and A. W. Van Wynsberghe, 2017. Characterization of Binding Pathways of Peramivir to H274Y Neuraminidase. Poster- 16th MERCURY Conference, Greenville, SC.
- Dacres, D. F.*, E. M. Lewis*, R. W. Wenner*, and A. W. Van Wynsberghe, 2017. Simulating the binding pathways of sialic acid and oseltamivir to H274Y neuraminidase with molecular dynamics solutions. Poster- 253rd ACS National Meeting, San Francisco, CA.
- Kang, G. M.* and A. W. Van Wynsberghe, 2017. Investigation of different binding kinetics among the neuraminidase inhibitors. Poster- 253rd ACS National Meeting, San Francisco, CA.
- Lewis, E. M.*, P. F. Marris*, L. M. Krause*, and A. W. Van Wynsberghe, 2017. Analysis of MM/GBSA free energy calculations to investigate the binding pathways of neuraminidase. Poster- 253rd ACS National Meeting, San Francisco, CA. (Awarded Computers in Chemistry Undergraduate Poster Award)
- Wenner, R. W.*, AB W. Abera*, B. J. Banman*, and A. W. Van Wynsberghe, 2017. Examining the binding pathways of peramivir to wild-type neuraminidase through molecular dynamics simulations and MM/GBSA analysis. Poster- 253rd ACS National Meeting, San Francisco, CA.
- Abera, AB W.*, B. J. Banman*, and A. W. Van Wynsberghe, 2016. Investigating the Binding pathways of Peramivir to Neuraminidase Through Molecular Dynamics
Simulations and MM/GBSA Analysis. Poster- 15th MERCURY Conference on Computational Chemistry, Lewisburg, PA. - Dacres, D. F.* E. M. Lewis*, R. W. Wenner*, J. F. Graziadei, and A. W. Van Wynsberghe, 2016. Simulating the Binding Pathways of Sialic Acid and Oseltamivir to H274Y Neuraminidase with Molecular Dynamics Simulations. Poster- 15th MERCURY Conference on Computational Chemistry, Lewisburg, PA.
- Lewis, E. M.*, P. F. Marris*, L. M. Krause, and A. W. Van Wynsberghe, 2016. Analysis of the Binding Pathways of Neuraminidase through MM/GBSA Post-Processing of
Equilibrium Molecular Dynamics Trajectories. Poster- 15th MERCURY Conference on Computational Chemistry, Lewisburg, PA. - Nash, K. L.* R. W. Wenner*, and A. W. Van Wynsberghe, 2016. Exploring the binding pathways of zanamivir to wild-type neuraminidase using molecular dynamics and MM/GBSA. Poster- 15th MERCURY Conference on Computational Chemistry, Lewisburg, PA.
- Dacres, D. F.* E. M. Lewis*, R. W. Wenner*, and A. W. Van Wynsberghe, 2016. Examining the Binding Pathways of Various Ligands to H274Y and Wild-Type Neuraminidase via Molecular Dynamics Simulations and MM/GBSA Analysis. Poster- 251st ACS National Meeting, San Diego, CA.
- Marris, P. F.* L. M. Krause*, J. Sørensen, and A. W. Van Wynsberghe, 2016. Investigating binding pathways to neuraminidase using MM/GBSA free energy analysis.
Poster- 251st ACS National Meeting, San Diego, CA. - Dacres, D. F.*, J. F. Graziadei*, P. F. Marris*, and A. W. Van Wynsberghe, 2015. Investigation of Sialic Acid Association Kinetics to H274Y Neuraminidase Using Molecular Dynamics Simulations. Poster- 14th MERCURY Conference on Computational Chemistry, Lewisburg, PA.
- Lewis, E. M.*, R. W. Wenner*, P. F. Marris*, and A. W. Van Wynsberghe, 2015. Examining the Binding Pathways of Oseltamivir to H274Y Neuraminidase. Poster- 14th MERCURY Conference on Computational Chemistry, Lewisburg, PA.
- Marris, P. F.*, L. M. Krause*, J. Sørensen, and A. W. Van Wynsberghe, 2015. Investigating binding pathways to neuraminidase using MM/GBSA free energy analysis. Poster- 14th MERCURY Conference on Computational Chemistry, Lewisburg, PA.
- Kang, G. M.*, D. J. Mermelstein*, R. B. Clayton*, and A. W. Van Wynsberghe, 2015. Investigation of different binding kinetics among the neuraminidase inhibitors. SciMix Poster Session- 249th ACS National Meeting, Denver, CO.
- Wenner, R. W.*, L. M. Krause*, J. F. Graziadei*, P. F. Marris*, and A. W. Van Wynsberghe, 2015. Calculation of the association trajectories of oseltamivir and sialic acid to wild type and H274Y viral neuraminidase. Poster- 249th ACS National Meeting, Denver, CO.
- Kang, G. M.*, D. J. Mermelstein*, R. B. Clayton*, and A. W. Van Wynsberghe, 2014. Investigation of Different Binding Kinetics Among the Neuraminidase Inhibitors. Poster- 13th MERCURY Conference on Computational Chemistry, Lewisburg, PA.
- Marris, P. F*, R. W. Wenner*, L. M. Krause*, J. Sørensen, A. R. Wu*, R. E. Amaro, and A. W. Van Wynsberghe, 2014. Investigating oseltamivir binding pathways to H274Y neuraminidase using molecular dynamics simulations and MM/GBSA analysis. Poster- 13th MERCURY Conference on Computational Chemistry, Lewisburg, PA.
- Wenner, R. W.*, P. F. Marris*, J. E. Adelman*, and A. W. Van Wynsberghe, 2014. Observing the Diffusion of Oseltamivir into the Active and Secondary Sites of Neuraminidase Wild Type and the H274Y Variant. Poster- 13th MERCURY Conference on Computational Chemistry, Lewisburg, PA.
- Alvarado, S. M.*, L. Krause*, A. R. Wu*, and A. W. Van Wynsberghe, 2013. Close-Range Behavior of Oseltamivir with Viral Neueraminidase. Poster- 12th MERCURY Conferencd on Computational Chemistry, Lewisburg, PA.
- Banman B. J.*, J. E. Adelman*, A. R. Wu*, and A. W. Van Wynsberghe, 2013. Observing the inter- and intramolecular Evants of Sialic Acid Binding to the Active Site of Neuraminidase. Poster- 12th MERCURY Conferencd on Computational Chemistry, Lewisburg, PA.
- Ho, J. H.*, C. M. Montangon*, R. B. Clayton*, and A. W. Van Wynsberghe, 2013. Investigation of the effects of molecular charges and water desolvation on the complex formation of Neuraminidase and its inhibitors. Poster- 12th MERCURY Conferencd on Computational Chemistry, Lewisburg, PA.
- O'Grady, C. E.*, P. Talpey*, and A. W. Van Wynsberghe, 2013. Development of a molecular docking exercise to elucidate principles of biophysical chemistry in the general chemistry lab. Poster- 12th MERCURY Conferencd on Computational Chemistry, Lewisburg, PA.
- Montagnon, C. M.*, R. B. Clayton*, and A. W. Van Wynsberghe, 2013. Investigation of the effects of electronic desolvation on oseltamivir binding kinetics to N1 influenza neuraminidase. Poster- 245th ACS National Meeting, New Orlenas, LA
- Wu, A. R.*, L. Krause*, and A. W. Van Wynsberghe, 2013. Elucidation of the molecular interactions between neuraminidase and sialic acid. Poster- 245th ACS National Meeting, New Orlenas, LA
UNDERGRADUATE STUDENT THESES SUPERVISED
-
Geum Mi Kang, Biochemistry, '17, "Investigation of Different Binding Kinetics Among the Neuraminidase Inhibitors and Sialic Acid."
-
Rich Wenner, Chemistry, '17, "A Computational Characterization of the Binding Kinetics of the Peramivir-Neuraminidase System."
-
Bryon Banman, Chemistry, '16, "Computationally Modeling Interactions Between Peramivir and Neuraminidase."
-
Jia Tsien Ho, Chemistry, '16, "Reinvestigating the Factors of Binding Kinetics in Oseltamivir and Sialic Acid."
-
Lizbeth DaBramo, Chemistry, '15, "The Secret in Soap: The Comparative Quantitative Analysis of Triclosan and Triclocarban."
-
Jordan F. Graziadei, Chemistry, '15, "Investigation of the Binding Kinetics of Sialic Acid to H275Y Influenza Neuraminidase."
-
Leah M. Krause, Chemistry, '14, "A Study of the Trajectories of Influenza Neuraminidase Binding using MM/GBSA Free Energy Calculations."
-
Daniel J. Mermelstein, Chemistry, '14, "Investigation of Oseltamivir Binding Kinetics to N1 Influenza Neuraminidase."
-
Clare E. O'Grady, Chemistry, '14, "The development and implementation of a bio-molecular docking exercise for the general and biochemistry laboratories."
-
Carmen M. Montagnon, Chemistry, '13, "Investigation of the Effects of Electronic Desolvation on Oseltamivir Binding Kinetics to N1 Influenza Neuraminidase."
-
Alvin R. Wu, Biochemistry, '13, "Investigating the Favored Binding Paths of Sialic Acid into the Active Site of Neuraminidase."
-
Erica L. Losito, Biohemistry, '12, "Role of Neuraminidase Electrostatics in the Binding of Ligands."
-
Laura D. Leonard, Chemistry, '11, "The Role of Secondary Sialic Acid Binding Sites on Avian and Human Influenza Neuraminidases."
-
Rebecaa S. Green, Chemistry, '11, "Identifying Favored Regions of Ligand Interaction Near the Active Site of Neuraminidase."
-
Carlos A. Rico, Chemical Physics, '10, "The Role of Secondary Sialic Acid Binding Sites on Avian Influenza Neuraminidases."
-
Thomas Morrell, Chemistry, '10, "The Impact of Short Term Dynamics on Ribonuclease A."
- Sam H. E. Cho, Chemistry, '10, "Conformation Dependence of the Thrombin-Thrombomodulin Interaction."
